| Ishita Mukerji
Associate Professor Ph.D. (Chemistry)
(860) 685-2422 |
We employ fluorescence and UV resonance Raman
spectroscopic methods for probing protein-nucleic acid and protein-protein
interactions. The Raman effect can be enhanced by several orders of magnitude
by exciting into or near to an absorption band. Thus, the excitation wavelength
can be used to probe different regions of the macromolecules. For example,
an excitation wavelength of 230 nm selectively investigates the aromatic
residues, Tyr and Trp; whereas, 260 nm selectively probes nucleic acid
residues. We exploit the resonance effect to separately investigate DNA
conformation from protein structure.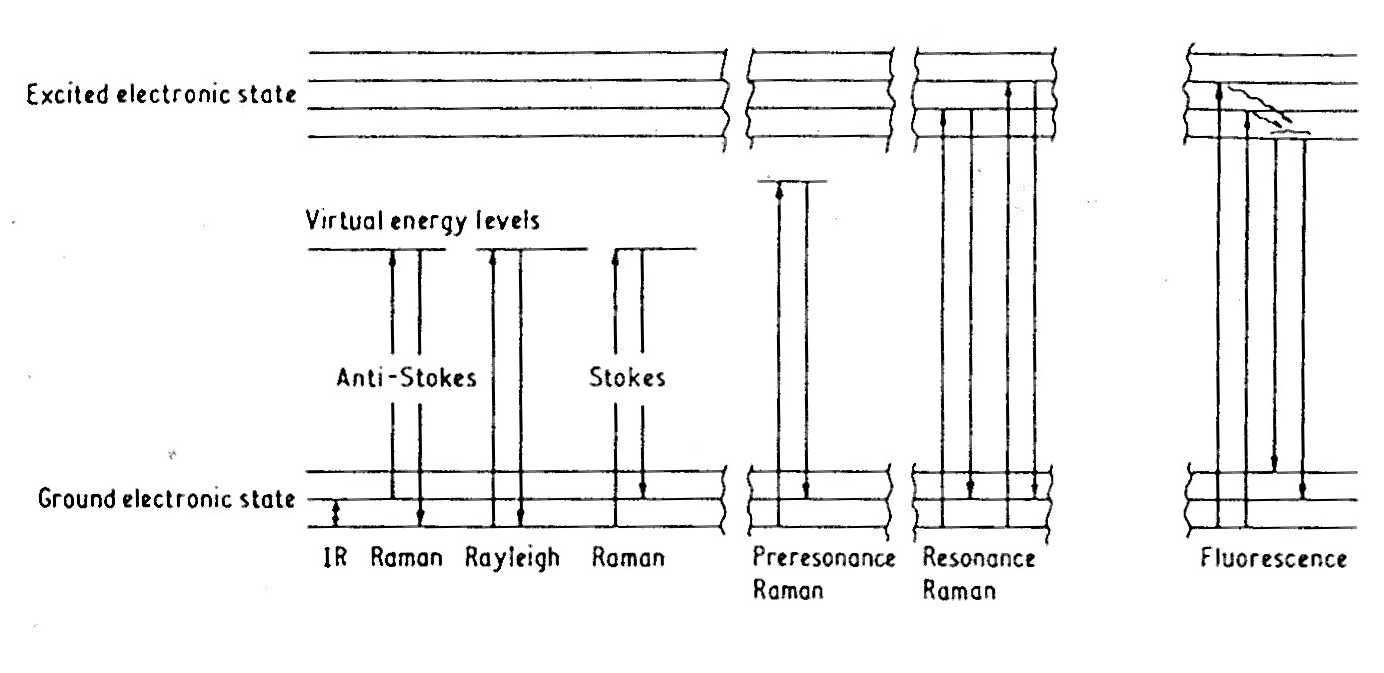
Protein-Nucleic Acid Interactions.Nucleotide-binding
proteins play an extremely important role as regulators of genomic function.
However, the molecular mechanism of these processes is not well understood,
since only a few crystal structures exist for protein-nucleic acid complexes.
We are addressing the mechanism of protein-mediated regulation of genetic
processes, such as repression, recombination or expression by investigating
the nucleotide-protein interface for a class of prokaryotic histone-like
proteins.
The stabilization of DNA in coil or loop structures is the postulated mechanism
by which these proteins participate in replication and inversion reactions
and also enhance binding of proteins such as Lac repressor and cAMP-activator
protein. It is this protein-induced deformation of DNA structure, which
in turn modulates its genetic function, that motivates our investigations.
We are studying the HU and IHF proteins from E.coli, which bind to the minor groove of DNA through two flexible b-strand regions. This type of interaction is of interest since the majority of previously characterized protein-nucleic acid interactions have typically involved direct contact between the protein a-helix and the major groove. The sequence specificity of the minor groove interaction is examined by monitoring H-bond pairings of nucleotide exocyclic amino and carbonyl groups. The vibrational modes of these exocyclic groups reflect the formation of H-bonds since molecular vibrations are dependent on the masses of the vibrating atoms, the molecular geometry, and the forces that restrain molecules in their equilibrium positions. We are also using fluorescence spectroscopy to probe the binding interaction to gain information regarding the global conformation of the protein-DNA complex. Our experiments focus on utilizing either the natural fluorophores in the protein (e.g. Tyr or Trp residues) or labeling the protein or the DNA with a fluorescent molecule. These fluorescence measurements allow us to probe the conformation of the DNA before and after it binds to the protein and fluorescence resonance energy transfer measurements reveal the relative proximity of the protein to the DNA.
Protein-Protein Interactions.Understanding
the forces that govern the interaction of proteins with one another assists
in the understanding of such processes as macromolecular assembly, chaperone-assisted
protein folding and protein translocation. We study the polymerization
of sickle cell hemoglobin as a paradigm for understanding protein-protein
interactions.  Polymerization
of sickle cell hemoglobin results from the one residue mutation (b6
Glu to Val) in the A helix of the protein. This one residue mutation creates
a hydrophobic surface that initiates the aggregation of the protein tetramers,
by interacting with the b85
Phe and b88 Leu
residues on an adjacent tetramer. Our studies are designed to investigate
the polymers as they are forming by monitoring the Phe Raman bands, which
are reflective of local environment. At present we have established that
the tertiary structure of Hb S tetramers differs from that of Hb A in the
region of the mutation. This tertiary structural change may have implications
for polymer formation. Our current work focuses on a Hb S derivative, which
allows us to chemically induce polymerization of the molecule. The mechanism
of polymerization is the same for the modified Hb S and native Hb S, as
shown by kinetic measurements. Electron micrographs of polymerized Hb demonstrate
that the fibers from the modified Hb S are similar in size and shape to
those formed by deoxy Hb S. Further work includes a study of Hb S fibers
at different stages in the polymerization process and a study of the effect
of anti-sickling drugs on Hb S fibers and tetramers.
Polymerization
of sickle cell hemoglobin results from the one residue mutation (b6
Glu to Val) in the A helix of the protein. This one residue mutation creates
a hydrophobic surface that initiates the aggregation of the protein tetramers,
by interacting with the b85
Phe and b88 Leu
residues on an adjacent tetramer. Our studies are designed to investigate
the polymers as they are forming by monitoring the Phe Raman bands, which
are reflective of local environment. At present we have established that
the tertiary structure of Hb S tetramers differs from that of Hb A in the
region of the mutation. This tertiary structural change may have implications
for polymer formation. Our current work focuses on a Hb S derivative, which
allows us to chemically induce polymerization of the molecule. The mechanism
of polymerization is the same for the modified Hb S and native Hb S, as
shown by kinetic measurements. Electron micrographs of polymerized Hb demonstrate
that the fibers from the modified Hb S are similar in size and shape to
those formed by deoxy Hb S. Further work includes a study of Hb S fibers
at different stages in the polymerization process and a study of the effect
of anti-sickling drugs on Hb S fibers and tetramers.
Selected Publications
- "A UV Resonance Raman Study of d(A+-G)10, a Single-Stranded Helix without Stacked or Paired Bases," I. Mukerji, M. C. Shiber, T. G. Spiro and J.R. Fresco, Biochemistry 34 (1995) 14300-14303.
- "Hemoglobin Allostery: Resonance Raman Spectroscopy of Kinetic Intermediates," V. Jayaraman, K. R. Rodgers, I. Mukerji and T. G. Spiro, Science 269 (1995) 1843-1848.
- "A UV Resonance Raman Study of Tetrads Formed in Hairpin Dimers of d(A-G)10 at Neutral pH," I. Mukerji, M. C. Shiber, T. G. Spiro and J. R. Fresco, Nucleic Acids Research 24 (1996) 5013-5020.
- "Temperature Dependent UV Resonance Raman Spectroscopy of the Premelting State of dA-dT DNA," S. S. Chan, R. H. Austin, I. Mukerji and T. G. Spiro, Biophys. J. 72 (1997) 1512-1520.
- "Conformational Changes in FmetHbS Probed with UV Resonance Raman and Fluorescence Spectroscopic Methods," L. Sokolov and I. Mukerji, J. Phys. Chem. 102 (1998) 8314-8319.
- "A UV Resonance Raman Investigation of Poly(rI): Evidence for Cation-Dependent Structural Perturbations," I. Mukerji, L. Sokolov and M.-R. Mihailescu, Biopolymers 46 (1998) 475-487.
- "New Light on Allostery: Dynamic Resonance Raman Spectroscopy of Hemoglobin Kempsey," X. Hu, K. R. Rodgers, I. Mukerji and T. G. Spiro, Biochemistry 38 (1999) 3462-3467.
- "Nucleic Acid Structure Investigated by UV Resonance Raman Spectroscopy: Protonation Effects and A-Tract Structure," L. Sokolov, K. Wojtuszewski, E. Tsukroff and I. Mukerji, J. Biomolec. Struct. Dyn. in press.
- "Solubility of Fluoromethemoglobin S: Effect of Phosphate and Temperature on Polymerization," M. E. Yohe, K. M. Sheffield and I. Mukerji, Biophys. J. in press
Lab Members
- Principal Investigator
- Ishita Mukerji
- Graduate Students
- Kelly Knee
- Iulia Vitoc
- Wendy Barber-Armstrong
- Andrew Moreno
- Undergraduates
- Maiko Kondo
- Russell Berg
TOP/BACK